 |
Web DSSR for dissecting the spatial structures of nucleic acids |
 |
| Home | Saturday, February 14, 2026 04:48 |
Tutorial
| Analysis | Reconstruction | Visualization |
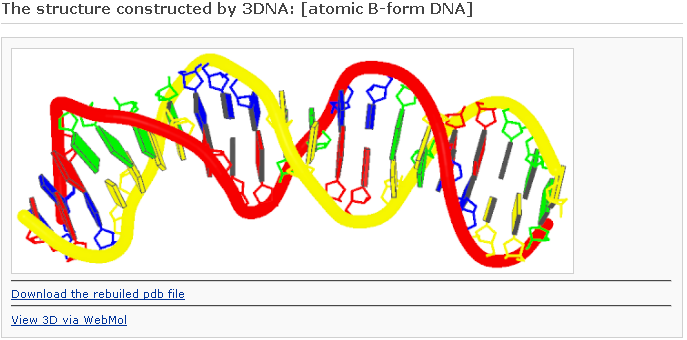
The reconstruction component allows the user to build three-dimensional models of arbitrary sequence and helical types, including 55 different fiber models and customized double-helical structures with base pairs and base-pair steps arranged according to user-supplied rigid-body parameters. The output includes a PDB-formatted file of the nucleic-acid base-pair and phosphorus atoms or an all-atom file with approximate sugar-phosphate backbones, and links for visualization via Webmol. The user can also construct models of DNA ‘decorated’ with proteins and other molecules by providing the requisite DNA-ligand structural templates and the locations of the binding sites.
Reconstruction of a fiber model
Fiber-model reconstruction is based on the repetition of an experimentally determined helical repeating unit. The user can select from 55 fiber models, with the three most popular ones (A-form, B-form, and C-form) placed at the top of the list of options. [Table of the 55 fiber models]
Example 2 -1: Choose a DNA fiber A-form (generic)
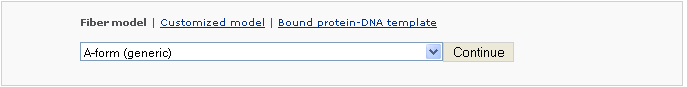
The generic fiber models accommodate arbitrary base sequences of any length. Note: only A, T, C, G characters can be entered as input sequence.
For a generic fiber, users have two options to input information:
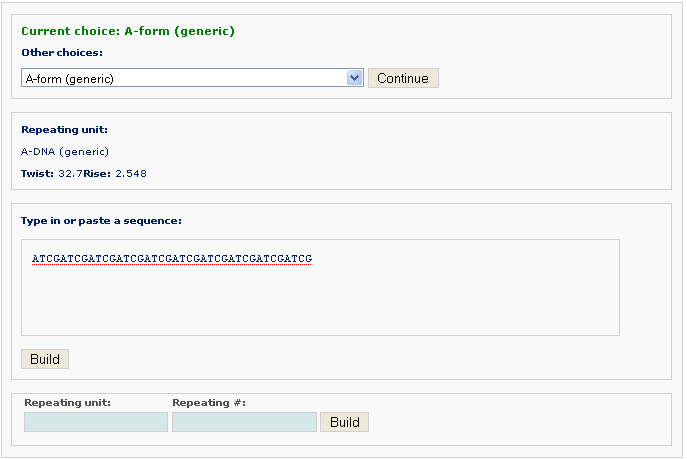
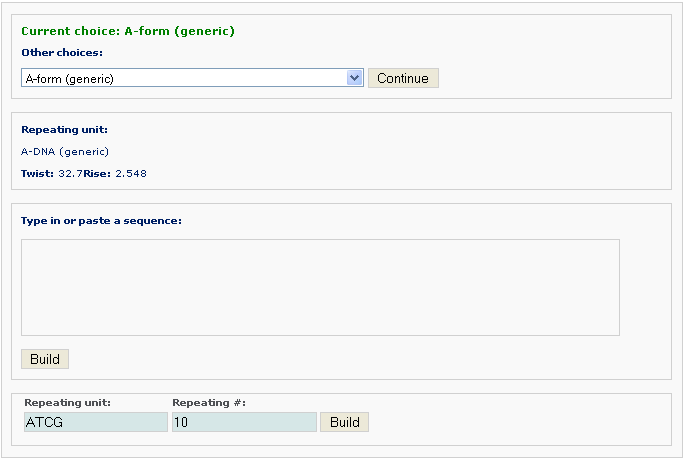
After clicking the ‘Build’ button, the user will be directed to the results page, which contains three sections: (1) a block representation of the reconstructed fiber model; [This option requires longer processing time] (2) a link to download the coordinates of the reconstructed model; (3) a link to view the atomic details of the reconstructed model.
One can click the “View 3D via Jmol” link to visualize the atomic details. Note: the Jmol* requires a Java Runtime Environment on your machine to work properly.
* Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org
One can click the “View 3D via WebMol” link to visualize the atomic details. Note: the WebMol** requires a Java Runtime Environment on your machine to work properly.
** Walther, D. (1997), WebMol: a Java-based PDB viewer. Trends Biochem. Sci., 22, 274-275.
Example 2 -2: Choose a DNA filber C-DNA poly d(GGT):poly d(ACC) (non-generic)
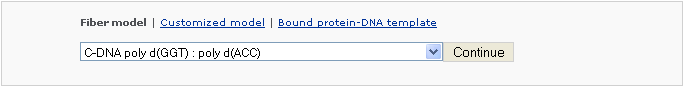
For non-generic fiber models, one must use the supplied nucleotide repeating unit. The length of the desired model is the product of the length of the repeating unit and the repeating # that the user enters.
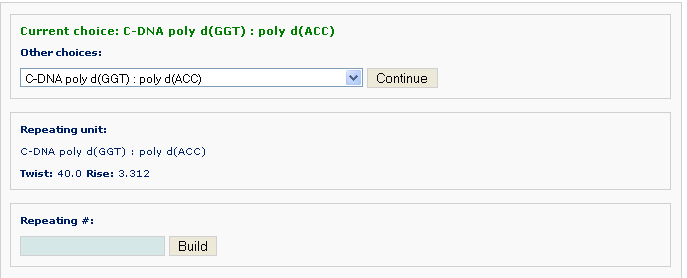
After clicking the ‘Build’ button, the user will be directed to a results page similar to that in Example 2 -1.
Reconstruction of a customized model
One can build a DNA model with customized base-pair step parameters or a mixture of different forms of DNA.
Example 2 -3: Choose a customized type Customized base-pair-step/nucleotide parameters
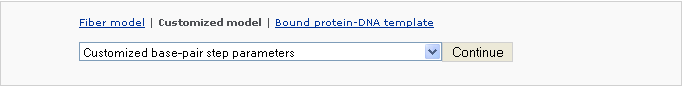
After clicking 'Continue' button, the user will be asked for uploading a base-pair step parameter file.
Either of the two testing files is illustrating the input format customized models (one for DNA, the other one for RNA). Please save it and upload to the w3DNA website for testing purpose). The customized format allows the user to specify the 3D arrangements of base pairs and base-pair steps in terms of rigid-body parameters in addition to the sequence and chain length. Non-canonical base pairs can be entered if the requisite base-parameters are given. Note that the base-pair step parameters are null for the first base pair.
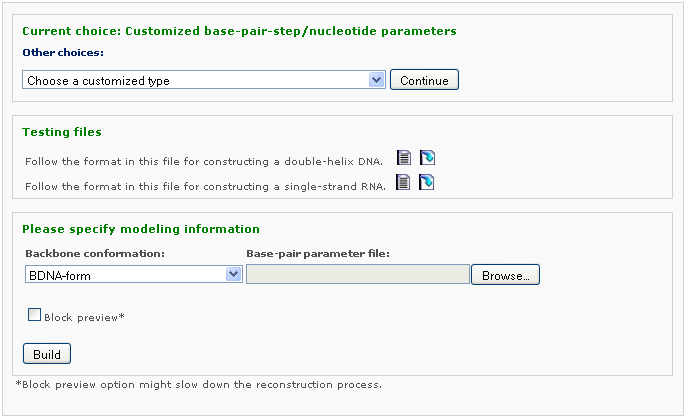
The user can specify the model type: (1) a full atomic model with an approximate backbone in PDB format, (2) a model containing only base and P atoms in PDB format. The user selects the type of rigid backbone that is introduced in the all-atom model: A-, B-, or C-type DNA.
After clicking “Build”, the user will be directed to a results page similar to that in Example 2-1.
Example 2 -4: Choose a customized type Combination of A-,B-, or C-form DNA
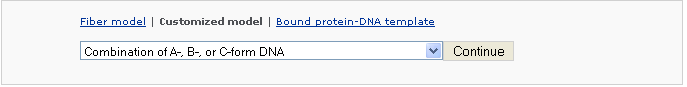
The user can also generate models that combine conformational segments of A-, B-, and C-DNA of arbitrary sequence and length.
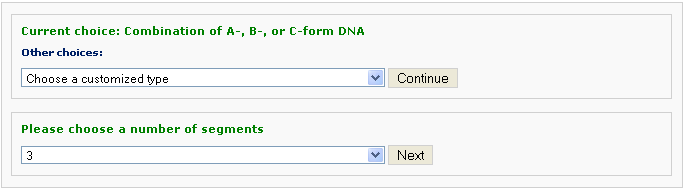
One chooses the number of different segments, each with a different conformational form, and the sequence of each segment. The sequence within each conformational segment can be specified in two ways, as an arbitrary string of bases or as a string of repeated base-paired units. The former information is entered with Option 1 and the latter with Option 2.
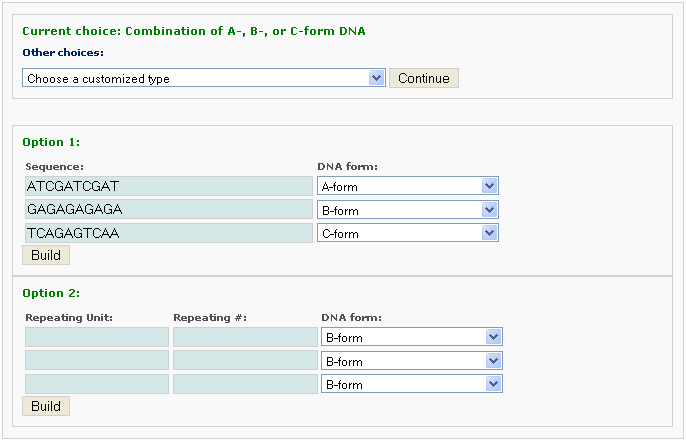
After clicking the ‘Build’ button, the user will be directed to a result page with information of the same form as Example 2-1.
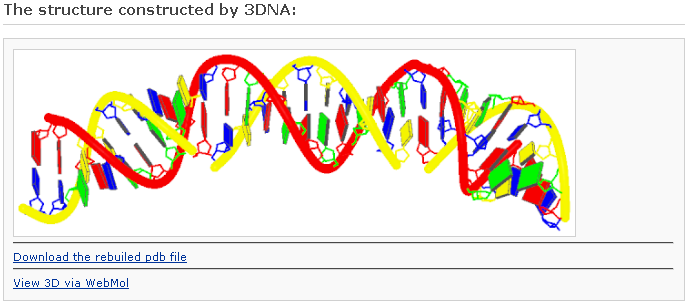
Reconstruction of a protein-bound DNA model
The user can generate models of DNA of arbitrary sequence and length ‘decorated’ with bound proteins and other ligands.
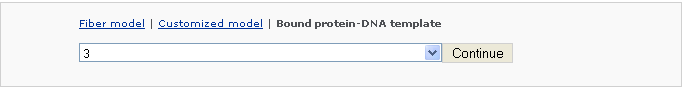
Example 2 -5: Choose a customized type Bound protein-DNA template
First the user selects the number of protein binding sites, say 3, then click the 'Continue' button.
The user is then asked to type in or paste the sequence of the DNA to be bound by proteins. The conformation of the unbound DNA can be specified as A-, B- or C-form. Please copy the following sequence to have a test:
Copy me:
ATCGTCGACC CGATTGACGC TAATCCGCGA ATCGTCGACC CGATTGACGC TAATCCGCGA ATCGTCGACC CGATTGACGC TAATCCGCGA ATGCTTCGCT
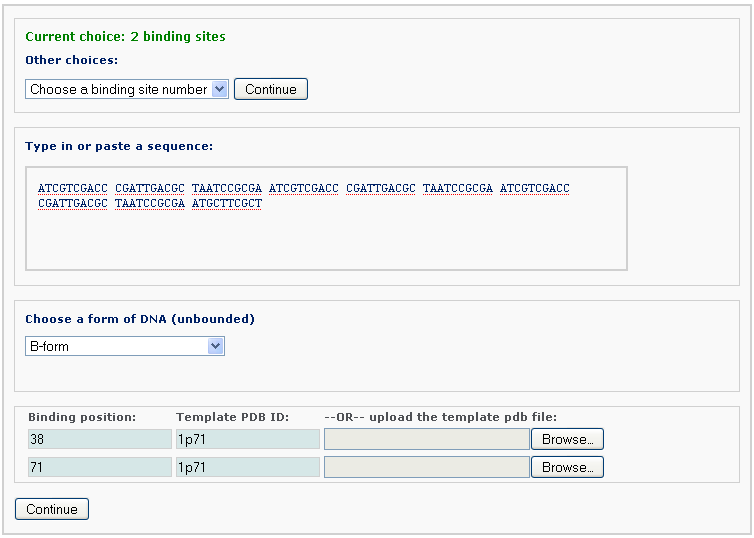
The user also supplies, for each binding site, the binding positions, which correspond to the locations of the central base or base-pair step of the specified protein-bound DNA structures. The template can be a PDB/NDB ID or an uploaded customized PDB-formatted structure. The computation will override the sequence in the PDB/NDB file, if a different sequence is entered.
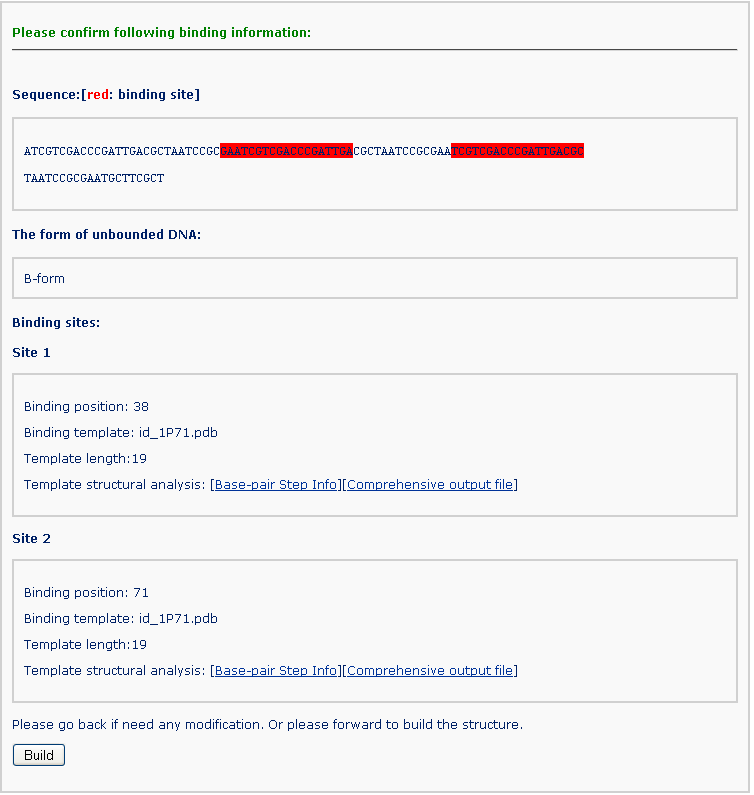
By clicking the ‘Continue’ button, the user is forwarded to a confirmation page, where the binding information can be verified. Warnings are given on this page for inappropriate binding input.
After clicking the ‘Build’ button, the user will be directed to a results page with information of the same form as Example 2-1, save for the addition of ribbon-like representations of the bound proteins. This block image will be displayed only when the 'Preview with block representation' was selected. [This option requires longer processing time]
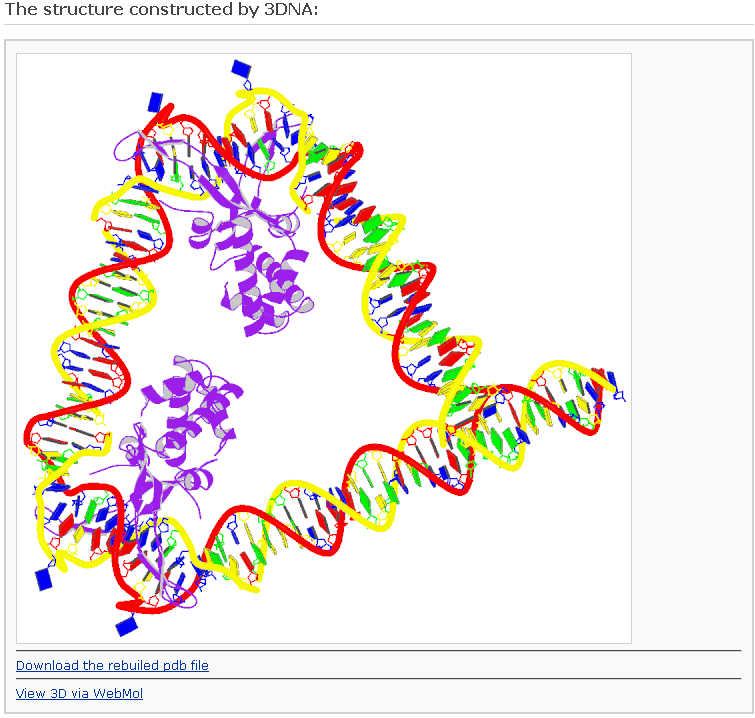
Table of regular DNA and RNA helical models
| Family | Molecule | Repeating sequence | Notes | Bases/turn | Strands | Reference |
| A | DNA | generic | 2 | 11 | (1,2) | |
| B | DNA | generic | 2 | 10 | (1,2) | |
| C | DNA | generic | 2 | 9.3 | (1,2) | |
| A | DNA | generic alternate | 2 | 11 | (3) | |
| B | DNA | generic alternate | 2 | 10 | (3) | |
| B | DNA | generic alternate | BI nucleotides | 2 | 10 | (4) |
| C | DNA | generic alternate | BII nucleotides | 2 | 9 | (4) |
| C | DNA | generic alternate | left-handed | 2 | 9.3 | (7) |
| A | DNA | ABr5U·ABr5U | 2 | 11 | (1,2) | |
| A | DNA | ATCGGAATGGT·TAGCCTTACCA | 2 | 11 | (1,2) | |
| B | DNA | CG·CG | 2 | 10 | (1,2) | |
| B | DNA | CCCCC·GGGGG | 2 | 10 | (1,2) | |
| B | DNA | A·T | Ca salt | 2 | 10 | (5) |
| B | DNA | A·T | Na salt | 2 | 10 | (5) |
| B | DNA | A·U | b | 2 | 10 | (1,2) |
| B´ | DNA | AATT·AATT | 2 | 10 | (1,2) | |
| B´ | DNA | A·T | alpha H DNA | 2 | 10 | (1,2) |
| B´ | DNA | A·T | beta1 | 2 | 10 | (1,2) |
| B´ | DNA | AI·CT | beta1 | 2 | 10 | (1,2) |
| B´ | DNA | A·T | beta2 H DNA beta | 2 | 10 | (1,2) |
| B´ | DNA | A·U | beta2 | 2 | 10 | (1,2) |
| B´ | DNA | AI·CT | beta2 | 2 | 10 | (1,2) |
| B* | DNA | A·T | high temperature | 2 | 11.4 | (6) |
| C | DNA | GGT·ACC | 2 | 9 | (1,2) | |
| C | DNA | GGT·ACC | 2 | 9 | (1,2) | |
| C | DNA | AG·CT | 2 | 9 | (1,2) | |
| C | DNA | AG·CT | 2 | 9 | (1,2) | |
| D | DNA | AAT·ATT | 2 | 8 | (1,2) | |
| D | DNA | CI·CI | 2 | 8 | (1,2) | |
| D | DNA | ATATAT·ATATAT | 2 | 8 | (1,2) | |
| D_A | DNA | AT·AT | 2 | 8.2 | (8) | |
| D_B | DNA | AT·AT | 2 | 8 | (8) | |
| L | DNA | GC·GC | 2 | (1,2) | ||
| S | DNA | CG·CG | CBGA, right-handed | 2 | 12 | (9) |
| S | DNA | GC·GC | CAGB, right-handed | 2 | 12 | (9) |
| Z | DNA | GC·GC | 2 | 12 | (1,2) | |
| Z | DNA | As4T·As4T | 2 | 14 | (1,2) | |
| DNA | C·I·C | 3 | 11(1,2) | |||
| DNA | T·A·T | 3 | 12(1,2) | |||
| DNA·RNA | A·dT | hybrid | 2 | 11(1,2) | ||
| DNA·RNA | dG·C | hybrid | 2 | 11.25(1,2) | ||
| DNA·RNA | dI·C | hybrid | 2 | 10(1,2) | ||
| DNA·RNA | dA·U | hybrid | 2 | 11(1,2) | ||
| A | RNA | A·U | 2 | 11 | (1,2) | |
| A | RNA | X·X | 2 | 11(1,2) | ||
| A | RNA | s2U·s2U | symmetric | 2 | 11(1,2) | |
| A | RNA | s2U·s2U | asymmetric | 2 | 11(1,2) | |
| A´ | RNA | I·C | 2 | 12 | (1,2) | |
| RNA | X·X | 2 | 10(1,2) | |||
| RNA | U·A·U | 3 | 11(1,2) | |||
| RNA | U·A·U | 3 | 11(1,2) | |||
| RNA | U·A·U | 3 | 12(1,2) | |||
| RNA | I·A·I | 3 | 12(1,2) | |||
| RNA | I·I·I·I | 4 | 11.5(1,2) | |||
| RNA | eC (O2´ ethyl) | 1 | 6(1,2) |
References
1. Chandrasekaran, R. and Arnott, S. (1989) In Saenger, W. (ed.), Landolt-Börnstein Numerical Data and Functional Relationships in Science and Technology, Group VII/1b, Nucleic Acids. Springer-Verlag, Berlin, pp. 31-170.
2. Arnott, S. (1999) In Neidle, S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, UK, pp. 1-38.
3. Premilat, S. and Albiser, G. (1983) Conformations of A-DNA and B-DNA in agreement with fiber X-ray and infrared dichroism. Nucleic Acids Res., 11, 1897-1908.
4. van Dam, L. and Levitt, M.H. (2000) BII nucleotides in the B and C forms of natural-sequence polymeric DNA: a new model for the C form of DNA with 40° helical twist. Journal of Molecular Biology, 304, 541-561.
5. Alexeev, D.G., Lipanov, A.A. and Skuratovskii, I.Y. (1987) The structure of poly(dA)·poly(dT) as revealed by an X-ray fibre diffraction. J. Biomol. Struct. Dynam., 4, 989-1011.
6. Premilat, S. and Albiser, G. (1997) X-ray fibre diffraction study of an elevated temperature structure of poly(dA)·poly(dT). Journal of Molecular Biology, 274, 64-71.
7. Premilat, S. and Albiser, G. (1984) Conformations of C-DNA in agreement with fiber X-ray and infrared dichroism. J. Biomol. Struct. Dynam., 2, 607-613.
8. Premilat, S. and Albiser, G. (2001) A new D-DNA form of poly(dA-dT)-poly(dA-dT): an A-DNA type structure with reversed Hoogsteen pairing. Eur. Biophys. J., 30, 404-410.
9. Premilat, S. and Albiser, G. (1999) Helix-helix transitions in DNA: fibre X-ray study of the particular cases poly(dG-dC) and poly(dA) 2poly(dT). Eur. Biophys. J., 28, 574-582.